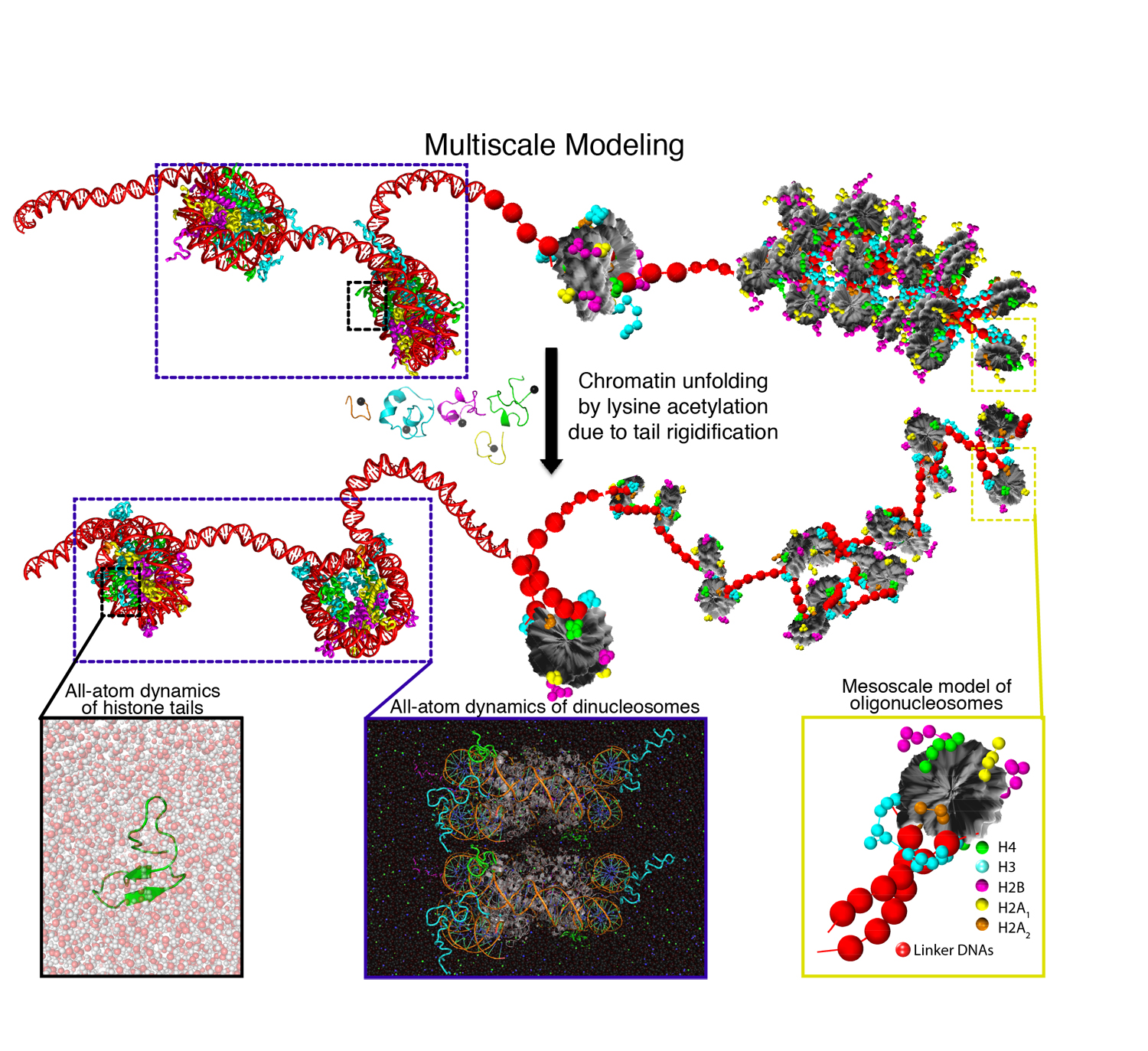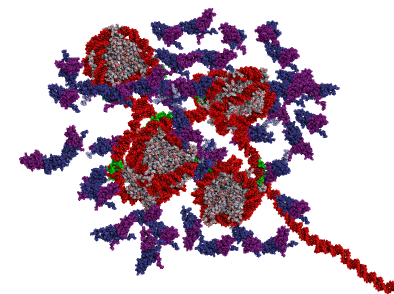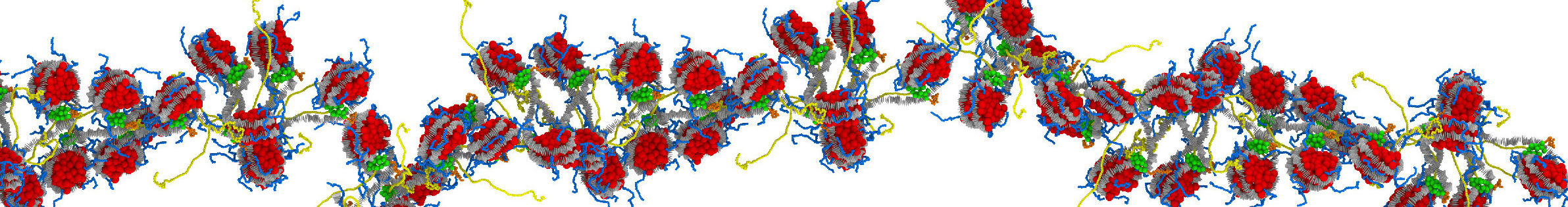
Our research aims at elucidating the molecular-level mechanisms behind control of 3D genome organisation at the nanoscale level. To do this, we develop and apply novel multiscale computational models for nanoscale chromatin that are anchored in all-atom molecular dynamics simulations, coarse-graining techniques, theory, and experiments from collaborators.
Stephen Farr
Multi-scale modelling of chromatin in functional domains
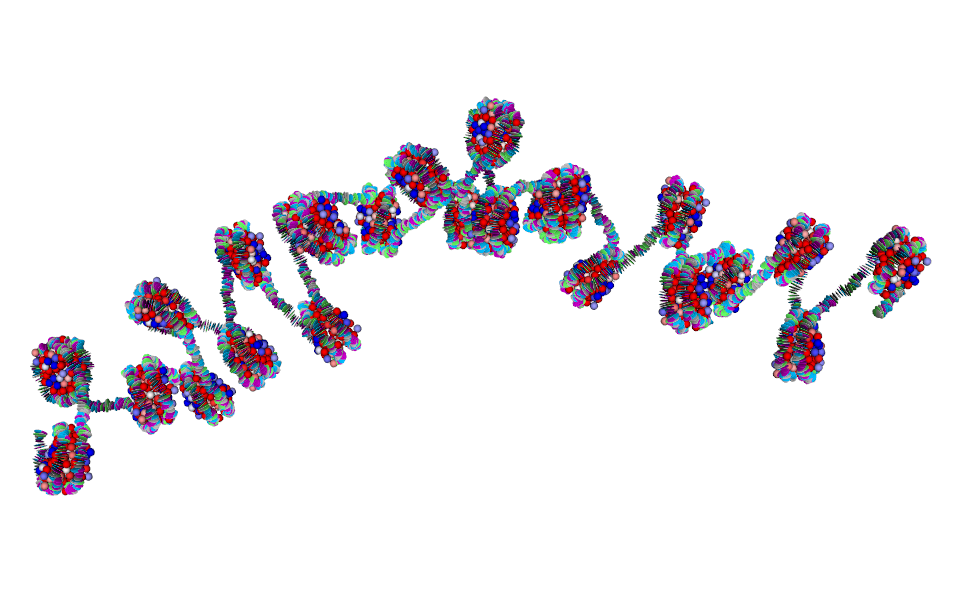
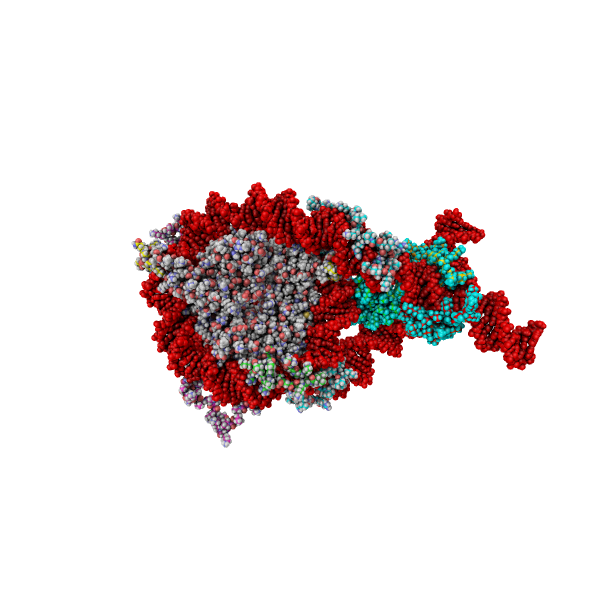
Steve's work integrates atomistic information of chromatin domains with two levels of coarse-grained Monte Carlo (MC) simulations and experimental data for validation. The high-resolution coarse-grained model part includes accurate molecular representations of nucleosomes with histone tails, epigenetic marks, architectural proteins bound to nucleosomes, protein oligomers, DNA unwrapping, and nucleosome sliding. The low-resolution coarse-grained model maps these factors implicitly to investigate nucleosome organization at Mb scales.
Akshay Sridhar
Epigenetic effects in chromatin structure
Adiran Garaizar
Liquid-liquid phase separation of chromatin domains
Adiran's work is in collaboration with the groups of Prof. Ernest Laue (Biochemistry, Cambridge) and Prof. Daan Frenkel (Chemistry, Cambridge). Adiran is developing a new coarse-grained model to investigate the phase behaviour of chromatin in conditions that lead to liquid demixing.
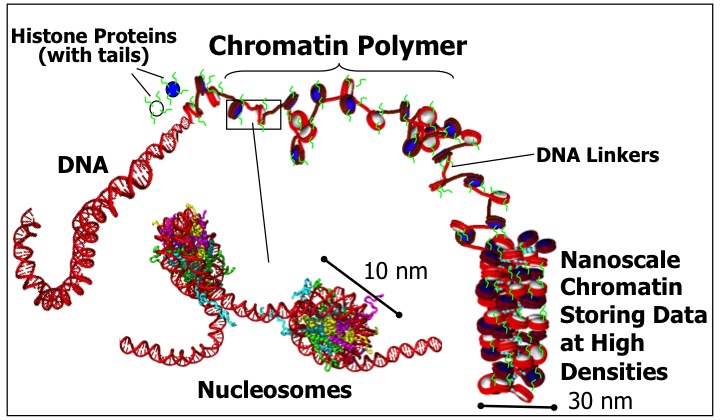
Chromatin Nanotechnology and Sustainability
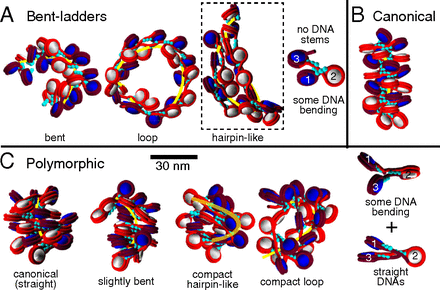
Structural Polymorphism